Aws Batch¶
This AMI is configured with the necessary software and daemons to run in AWS Batch. Part of the application exploits FPGAs to speedup the computation of the heaviest workloads. These devices needs to be visibile to the Docker container that will be employed. Furthermore, to exploit the accelerated version of the software provided by Huxelerate, it is necessary to mount specific volumes on the Docker container.
Volume Mapping and Mount Points¶
Here follows an example of volumes and mount points in a job definition. It is important to share with the Docker container the folder where the script and the libraries to handle the FPGA are installed, and the binaries provided by Huxelerate.
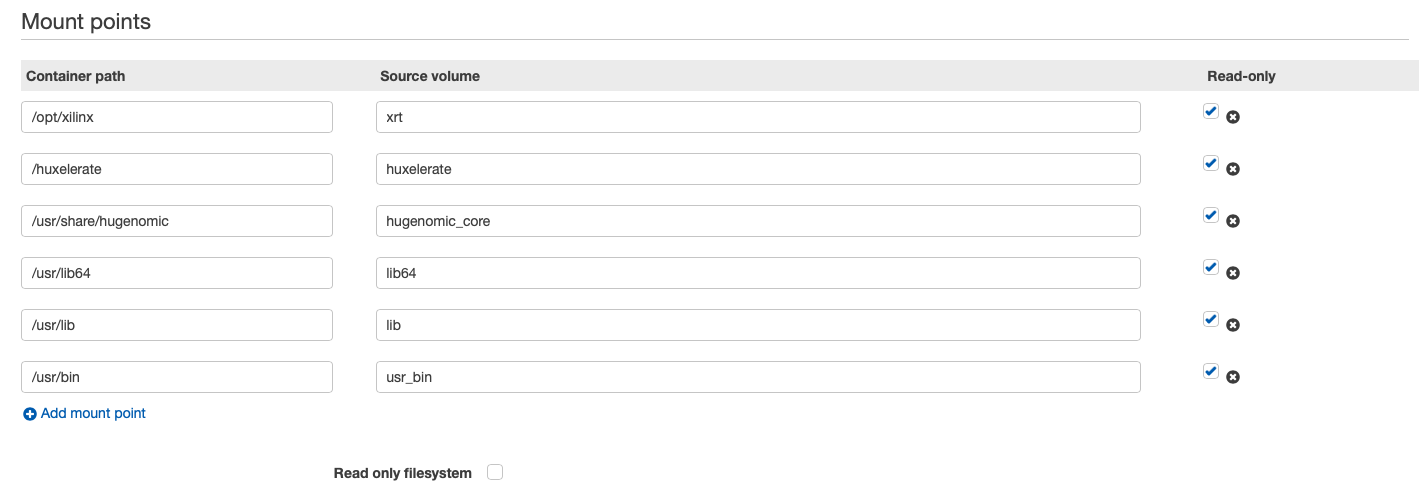
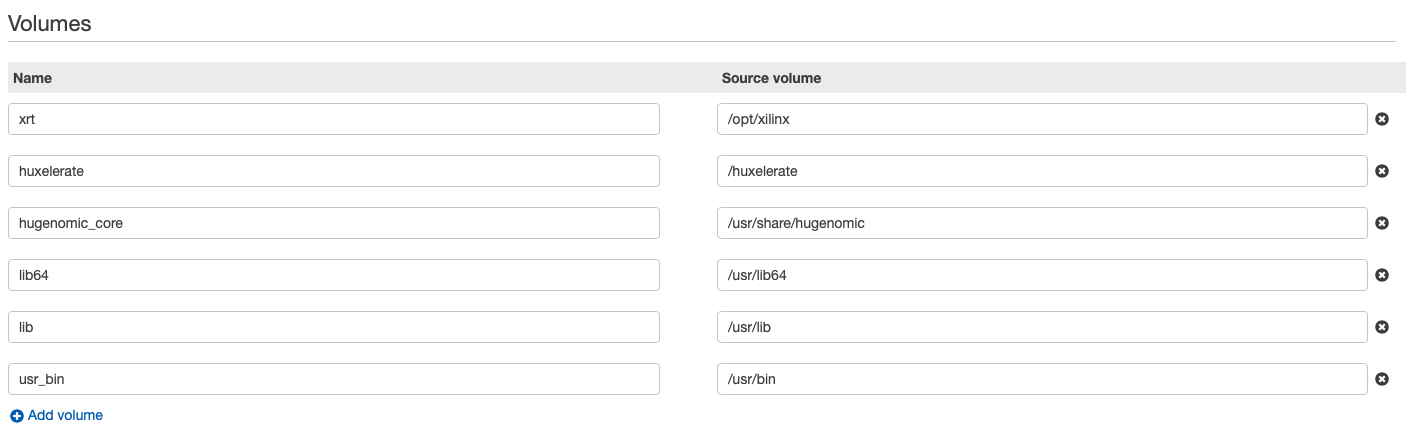
Docker requires privileged run to access the FPGAs, in the job definition remember to specify it:

The AMI provides the binaries from Hugenomic Nanopolish and minimap2, you are responsible to install any other software needed and to setup the environment using the script provided by Xilinx.
source /opt/xilinx/xrt/setup.sh
Once the environment is configured, the binaries are available in the folder:
/huxelerate/hugenomic/
# run nanopolish as
/huxelerate/hugenomic/hug-nanopolish.exe
Note
The default number of threads for module eventalign is 12 and it is optimized to be used on the f1.2xlarge instance. It is however possible to overload the number of threads by using the -t option.
Note
When using the eventalign module with –samples, the output produced can be massive. To improve the performance of the AMI, it is suggested to use two different volumes, one for reading the inputs and one for storing the outputs of the computation.
Example Docker Images¶
We provide three base Docker images, accessible from Huxelerate Docker Hub.
nanopolish_base is a Centos7 image with basic useful tools that could be necessary when running nanopolish (Development Tools, git, wget, tar and zlib).
fetch_and_run is a Centos7 image that can be exploited as in the example described in the Fetch and Run example by AWS.
fetch_and_run_hug_nanopolish_example is based on the previous image, plus minimap2 and samtools 1.10 installed in the /softwares folder.
Using the fetch_and_run images, it is possible to upload a bash script to S3, specify it as an environment variable and launch the computation. The container will automatically download and execute the script.
Note
To access S3 from the docker container, you will have to create a specific IAM role with “AmazonS3ReadOnlyAccess” permission.
Here is an example script and job environmental variables definition that can be used with the fetch_and_run docker container.
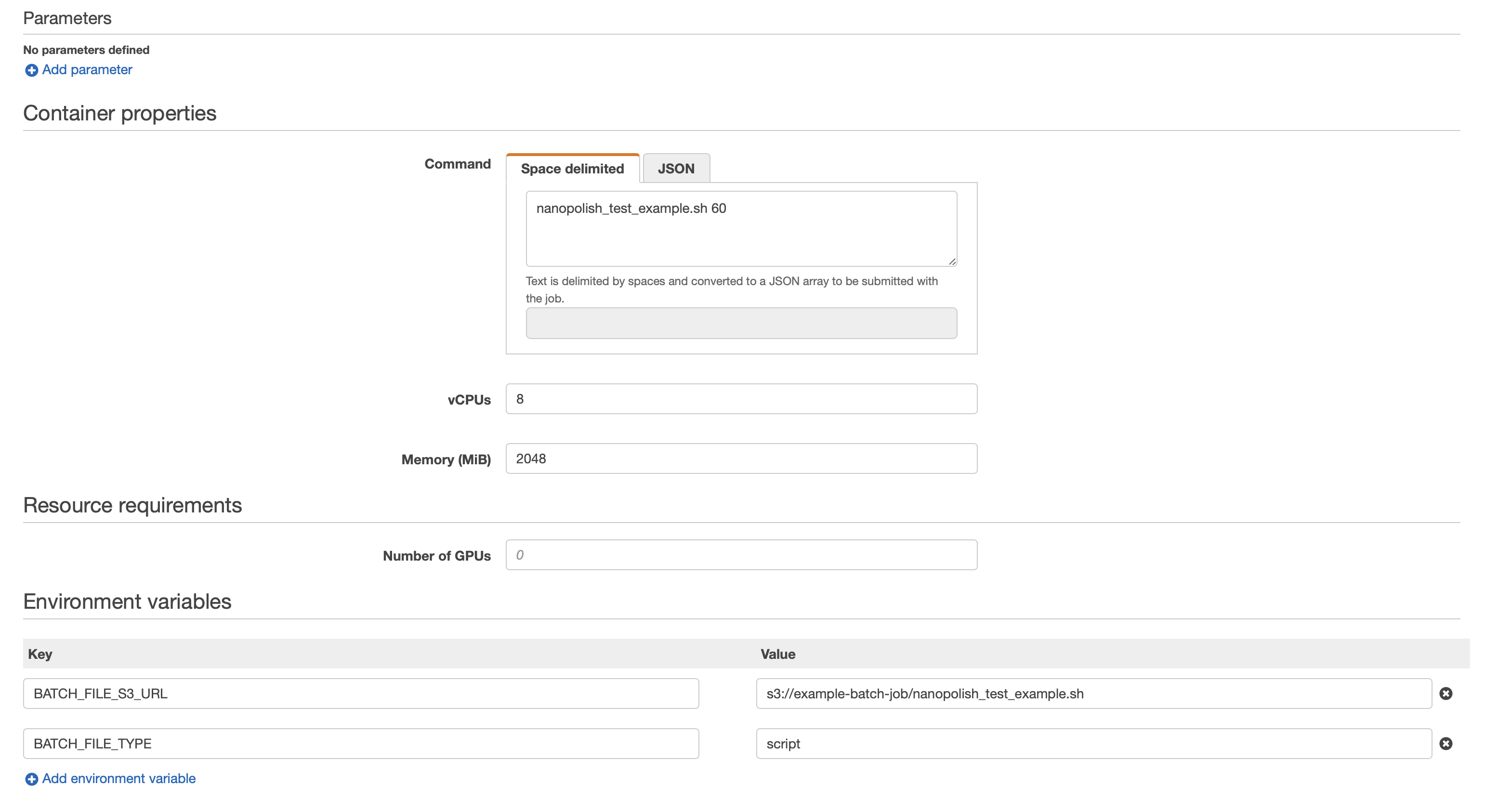
The BATCH_FILE_S3_URL variabile containes the S3 address of the script, while the BATCH_FILE_TYPE needs to be set to script.
Note
The docker images are provided just as examples and comes with no warranty.
Script to run ecoli example¶
Here you can find an example script that can be uploaded to S3 to perform an alignment example. To test the script, it is possible to use the fetch_and_run_hug_nanopolish_example image.
# source Xilinx script to setup the environment
source /opt/xilinx/xrt/setup.sh
# download and test an example dataset
wget http://s3.climb.ac.uk/nanopolish_tutorial/ecoli_2kb_region.tar.gz
tar -xvf ecoli_2kb_region.tar.gz
cd ecoli_2kb_region
curl -o ref.fa https://ftp.ncbi.nih.gov/genomes/archive/old_genbank/Bacteria/Escherichia_coli_K_12_substr__MG1655_uid225/U00096.ffn
# use hug-nanopolish executable to perform the computation
/huxelerate/hugenomic/hug-nanopolish.exe index -d fast5_files/ reads.fasta
/huxelerate/hugenomic/minimap2 -ax map-ont -t 8 ref.fa reads.fasta | /softwares/samtools-1.10/samtools sort -o reads-ref.sorted.bam -T reads.tmp
/softwares/samtools-1.10/samtools index reads-ref.sorted.bam
/huxelerate/hugenomic/hug-nanopolish.exe eventalign \
--reads reads.fasta \
--bam reads-ref.sorted.bam \
--genome ref.fa \
--scale-events > reads-ref.eventalign.txt
In this particular example, the script supposes that the Docker container contains the version 1.10 of samtools installed in the /softwares folder (as in the fetch_and_run_hug_nanopolish_example Docker image).